第 4 章 进阶技巧
程序的可读性和执行效率是两个很重要的要求.这一章主要介绍如何提高这两点.
4.1 apply函数族
apply并不能提高执行效率,只能使代码简洁易读.
详细的介绍可以在apply函数族介绍查看,其中给了一些简单的例子.下面演示一下稍微复杂的用法.
对矩阵按列进行操作,每列的奇数项求和,偶数项求和:
m <- matrix(1:12,4,3)
m## [,1] [,2] [,3]
## [1,] 1 5 9
## [2,] 2 6 10
## [3,] 3 7 11
## [4,] 4 8 12apply(m, 2, tapply,rep(1:2,2), sum)## [,1] [,2] [,3]
## 1 4 12 20
## 2 6 14 22对矩阵按列进行操作,每列前两项求和,后两项求和:
apply(m, 2, tapply,rep(1:2,each=2), sum)## [,1] [,2] [,3]
## 1 3 11 19
## 2 7 15 23下面看一个3维情况的例子.
a <- array(1:24,dim = c(2,3,4))
a## , , 1
##
## [,1] [,2] [,3]
## [1,] 1 3 5
## [2,] 2 4 6
##
## , , 2
##
## [,1] [,2] [,3]
## [1,] 7 9 11
## [2,] 8 10 12
##
## , , 3
##
## [,1] [,2] [,3]
## [1,] 13 15 17
## [2,] 14 16 18
##
## , , 4
##
## [,1] [,2] [,3]
## [1,] 19 21 23
## [2,] 20 22 24固定1个维度,对另外2个维度求和:
apply(a,1,sum)## [1] 144 156这样看起来不太直观,我们利用aperm函数调整数组的维度顺序:
aperm(a,c(2,3,1))## , , 1
##
## [,1] [,2] [,3] [,4]
## [1,] 1 7 13 19
## [2,] 3 9 15 21
## [3,] 5 11 17 23
##
## , , 2
##
## [,1] [,2] [,3] [,4]
## [1,] 2 8 14 20
## [2,] 4 10 16 22
## [3,] 6 12 18 24固定2个维度,对1个维度求和:
apply(a,c(2,3),sum)## [,1] [,2] [,3] [,4]
## [1,] 3 15 27 39
## [2,] 7 19 31 43
## [3,] 11 23 35 47固定2个维度,对1个维度分组求和:
apply(a,c(1,2),tapply,rep(c(-1,-2),2), sum)## , , 1
##
## [,1] [,2]
## -2 26 28
## -1 14 16
##
## , , 2
##
## [,1] [,2]
## -2 30 32
## -1 18 20
##
## , , 3
##
## [,1] [,2]
## -2 34 36
## -1 22 24对第三个维度奇数项、偶数项分别求和(奇数项是-1组,偶数项是-2组).
4.2 并行
单次模拟时间越长,重复次数越多,并行得到的提升越明显.
R中有很多并行包,可以在 不同并行包比较查看对比.
这里介绍的foreach是比较友好的一个包.
下面以线性模型估计系数为例,给出示例代码.
sim_single可以在 单次估计程序 查看.其中SLP参数用于增加单次模拟的计算量(运行时间).
4.2.1 低重复次数,低计算量
source("code/parallel-demo.R")
library("foreach")
library("doParallel")
beta0 = c(1,-2,3)
N = c(50,100,200)
distribution= c(rnorm,rcauchy)
SIM = 500
tstart=Sys.time()
cl <- makeCluster(detectCores())
registerDoParallel(cl)
result <- foreach(n=N,.combine = rbind) %:%
foreach(dst=distribution,.combine = c) %:%
foreach(i=1:SIM,.combine = '+',
.packages = c("MASS") )%dopar%{
sim_single(n,beta0,SLP=FALSE,mydist = dst)
}
stopImplicitCluster()
t.end=Sys.time()
t.end-tstart## Time difference of 2.182 secsresult/SIM## [,1] [,2] [,3] [,4] [,5] [,6]
## result.1 0.9988 -2.006 3.002 -0.05712 -1.969 1.962
## result.2 0.9979 -1.996 2.997 2.41988 -4.019 1.518
## result.3 0.9985 -2.002 3.007 0.62494 -2.432 2.903把申请cluster的命令去掉,%dopar%换成%do%程序就会按串行执行
source("code/parallel-demo.R")
library("foreach")
library("doParallel")
beta0 = c(1,-2,3)
N = c(50,100,200)
distribution= c(rnorm,rcauchy)
SIM = 500
tstart=Sys.time()
#cl <- makeCluster(detectCores())
#registerDoParallel(cl)
result <- foreach(n=N,.combine = rbind) %:%
foreach(dst=distribution,.combine = c) %:%
foreach(i=1:SIM,.combine = '+',
.packages = c("MASS") )%do%{
sim_single(n,beta0,SLP=FALSE,mydist = dst)
}
#stopImplicitCluster()
t.end=Sys.time()
t.end-tstart## Time difference of 2.285 secsresult/SIM## [,1] [,2] [,3] [,4] [,5] [,6]
## result.1 1.0005 -1.997 3.003 7.4661 -4.5211 -0.2274
## result.2 0.9939 -2.002 3.003 -0.8143 -0.4892 4.1754
## result.3 1.0036 -2.002 2.996 0.4728 -1.9347 0.97974.2.2 高重复次数,低计算量
增加到1000次.
并行:
source("/Users/wang/Documents/GitHub/Simulation-in-R/code/parallel-demo.R")
library("foreach")
library("doParallel")
beta0 = c(1,-2,3)
N = c(50,100,200)
distribution= c(rnorm,rcauchy)
SIM = 1000
tstart=Sys.time()
cl <- makeCluster(detectCores())
registerDoParallel(cl)
result <- foreach(n=N,.combine = rbind) %:%
foreach(dst=distribution,.combine = c) %:%
foreach(i=1:SIM,.combine = '+',
.packages = c("MASS") )%dopar%{
sim_single(n,beta0,SLP=FALSE,mydist = dst)
}
stopImplicitCluster()
t.end=Sys.time()
t.end-tstart## Time difference of 3.403 secsresult/SIM## [,1] [,2] [,3] [,4] [,5] [,6]
## result.1 1.007 -2.013 2.999 0.1556 -2.407 3.176
## result.2 1.008 -2.002 2.996 0.7327 -2.565 3.577
## result.3 1.000 -2.000 2.997 2.5792 -2.113 2.966串行:
source("code/parallel-demo.R")
library("foreach")
library("doParallel")
beta0 = c(1,-2,3)
N = c(50,100,200)
distribution= c(rnorm,rcauchy)
SIM = 1000
tstart=Sys.time()
#cl <- makeCluster(detectCores())
#registerDoParallel(cl)
result <- foreach(n=N,.combine = rbind) %:%
foreach(dst=distribution,.combine = c) %:%
foreach(i=1:SIM,.combine = '+',
.packages = c("MASS") )%do%{
sim_single(n,beta0,SLP=FALSE,mydist = dst)
}
#stopImplicitCluster()
t.end=Sys.time()
t.end-tstart## Time difference of 4.604 secsresult/SIM## [,1] [,2] [,3] [,4] [,5] [,6]
## result.1 1.0017 -1.999 3.000 1.5062 -2.2061 2.577
## result.2 1.0032 -1.990 2.997 3.9836 -0.5012 3.743
## result.3 0.9991 -1.997 2.997 0.8568 -2.0615 2.7184.2.3 低重复次数,高计算量
增加每次模拟的计算量,延长单次模拟的时间.
并行:
source("/Users/wang/Documents/GitHub/Simulation-in-R/code/parallel-demo.R")
library("foreach")
library("doParallel")
beta0 = c(1,-2,3)
N = c(50,100,200)
distribution= c(rnorm,rcauchy)
SIM = 500
tstart=Sys.time()
cl <- makeCluster(detectCores())
registerDoParallel(cl)
result <- foreach(n=N,.combine = rbind) %:%
foreach(dst=distribution,.combine = c) %:%
foreach(i=1:SIM,.combine = '+',
.packages = c("MASS") )%dopar%{
sim_single(n,beta0,SLP=TRUE,mydist = dst)
}
stopImplicitCluster()
t.end=Sys.time()
t.end-tstart## Time difference of 9.387 secsresult/SIM## [,1] [,2] [,3] [,4] [,5] [,6]
## result.1 1.0101 -2.001 2.994 1.373 -1.572 2.844
## result.2 0.9968 -2.013 2.992 3.813 -4.572 8.934
## result.3 0.9996 -1.997 3.000 7.350 -4.860 3.662串行:
source("code/parallel-demo.R")
library("foreach")
library("doParallel")
beta0 = c(1,-2,3)
N = c(50,100,200)
distribution= c(rnorm,rcauchy)
SIM = 500
tstart=Sys.time()
#cl <- makeCluster(detectCores())
#registerDoParallel(cl)
result <- foreach(n=N,.combine = rbind) %:%
foreach(dst=distribution,.combine = c) %:%
foreach(i=1:SIM,.combine = '+',
.packages = c("MASS") )%do%{
sim_single(n,beta0,SLP=TRUE,mydist = dst)
}
#stopImplicitCluster()
t.end=Sys.time()
t.end-tstart## Time difference of 38.65 secsresult/SIM## [,1] [,2] [,3] [,4] [,5] [,6]
## result.1 1.0086 -2.010 2.998 12.4454 -0.4945 0.05709
## result.2 0.9932 -1.998 2.997 -0.6948 -3.1508 1.68111
## result.3 0.9986 -2.003 3.001 0.8107 -1.7691 3.496744.2.4 模型选择
除了蒙特卡洛模拟之外,另外一个很适合并行的应用是模型选择.下面的例子用AIC对线性模型进行选择:
AIClm <- function(Y,X,ind){#单次计算AIC,ind为纳入模型的变量下标
AIC(lm(Y~X[,ind]))
}
gen_ind <- function(bt, index) bt*index#生成候选模型下标
n = 100
p = 18
m0 = p%/%3
p0 = sample(1:p,m0)
beta = rep(0,p)
beta[p0] = 1:2
X <- matrix(rnorm(n*p),n,p)
Y <- X%*%beta + rnorm(n)
##生成所有候选模型的下标
pl <- 1:(2^p - 1)
mt <- matrix(0,2^p-1,p)
for (i in pl) {
mt[i,]=rev(as.integer(intToBits(i)[1:(p)] ))
}
index = 1:p
lst = t(apply(mt, 1, gen_ind,index=index))
#并行
library("foreach")
library("doParallel")
tstart=Sys.time()
cl <- makeCluster(detectCores())
registerDoParallel(cl)
foreach(i=1:(2^p-1),.combine = c) %dopar%{
AIClm(Y,X,lst[i,])
}->result
stopImplicitCluster()
t.end=Sys.time()
t.end-tstart## Time difference of 1.405 minswhich(lst[which.min(result),]!=0)## [1] 1 5 6 8 11 12 13 15 16 17 18which(beta!=0)## [1] 6 8 13 16 17 18#串行
tstart=Sys.time()
cl <- makeCluster(detectCores())
registerDoParallel(cl)
foreach(i=1:(2^p-1),.combine = c) %do%{
AIClm(Y,X,lst[i,])
}->result
stopImplicitCluster()
t.end=Sys.time()
t.end-tstart## Time difference of 3.495 minswhich(lst[which.min(result),]!=0)## [1] 1 5 6 8 11 12 13 15 16 17 18which(beta!=0)## [1] 6 8 13 16 17 184.3 Rcpp
Rcpp提供了R与C++的无缝接口,可以很方便的在R中调用编写的C++程序.
可以通过cppFunction直接在R中编写:
Rcpp::cppFunction('int add(int x, int y, int z) {
int sum = x + y + z;
return sum;
}')
add## function (x, y, z)
## .Call(<pointer: 0x10af678f0>, x, y, z)add(1, 2, 3)## [1] 6但是这种方式在编写(没有语法高亮)、调试(定位编译报错行号)时都有不便.推荐单独编写cpp文件,通过sourceCpp加载.
下面以矩阵按列求和为例,比较col_mean(自己编写的C++函数)11、colMeans(R base中提供的注重速度的函数)、mean_R(在R中用编写的函数)以及apply的运行速度.
Rcpp::sourceCpp('code/Rcpp-demo.cpp')##
## > mean_R <- function(X) {
## + n = dim(X)[1]
## + m = dim(X)[2]
## + cm <- rep(0, m)
## + for (i in 1:n) {
## + cm = cm + X[i, ]
## + }
## + .... [TRUNCATED]m = 200
n = 100
X <- matrix(rnorm(m*n),m,n)
col_mean(X) -> l1
mean_R(X) -> l2
all.equal(l1,l2)## [1] TRUEcolMeans(X) -> l3
all.equal(l1,l3)## [1] TRUEapply(X, 2, mean) -> l4
all.equal(l1,l4)## [1] TRUEbench::mark(
col_mean(X),
colMeans(X),
mean_R(X),
apply(X, 2, mean),
check = FALSE,relative = TRUE
)->results
ggplot2::autoplot(results)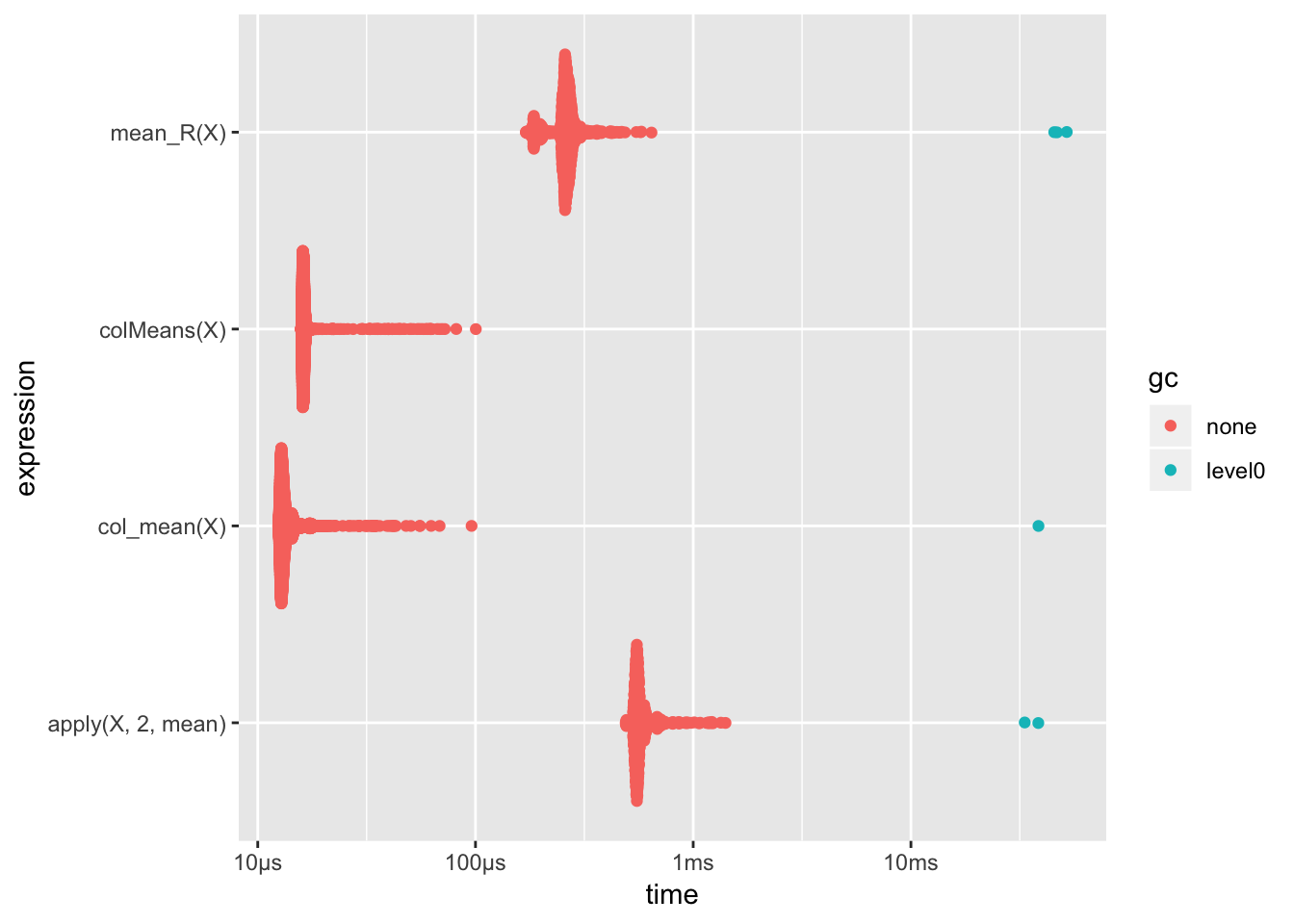
对比结果如图所示,其中gc12是一个关于内存使用的指标,越低越好.
apply和在R中用循环编写函数速度差不多,用C++编写的函数明显比其他快,甚至比base库中的函数还要快.不过,当我们增加矩阵的大小时,就会发现不一样的结果:
Rcpp::sourceCpp('code/Rcpp-demo.cpp')##
## > mean_R <- function(X) {
## + n = dim(X)[1]
## + m = dim(X)[2]
## + cm <- rep(0, m)
## + for (i in 1:n) {
## + cm = cm + X[i, ]
## + }
## + .... [TRUNCATED]m = 2000
n = 1000
X <- matrix(rnorm(m*n),m,n)
col_mean(X) -> l1
mean_R(X) -> l2
all.equal(l1,l2)## [1] TRUEcolMeans(X) -> l3
all.equal(l1,l3)## [1] TRUEapply(X, 2, mean) -> l4
all.equal(l1,l4)## [1] TRUEbench::mark(
col_mean(X),
colMeans(X),
mean_R(X),
apply(X, 2, mean),
check = FALSE,relative = TRUE
)->results
ggplot2::autoplot(results)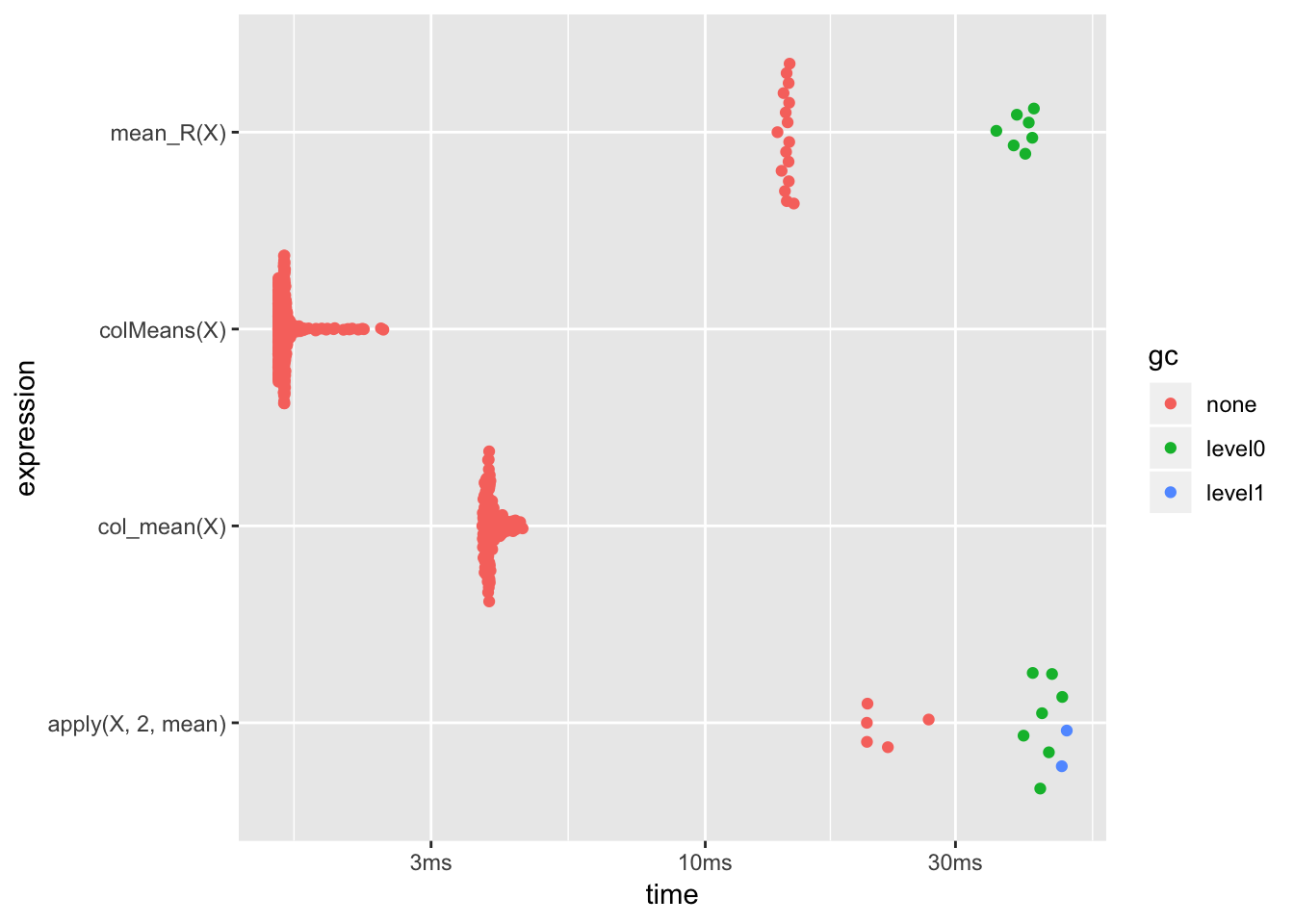
可以看到,还是base库中提供的函数速度最快.
4.3.1 RcppParallel
C++并行库
这里提供一个RcppParallel的例子: 核估计
下面是三个关于线性代数的库,https://gist.github.com/wolfv/ca3ac2b24e1daf70f85eac18ec7b1b8f 这个例子的测试结果表明xtensor最快
4.3.2 RcppArmadillo
线性代数库 官方文档
4.3.3 RcppEigen
线性代数库(更快一点,但是不友好) 官方文档